


The often-quoted "central dogma of molecular biology" states that the information stored in DNA is translated into functional proteins and that RNA, which acts as a messenger between the genome and the translation machinery, faithfully reflects the original code of its DNA template. This rule is suspended by RNA editing which alters RNA sequences at specific sites (Sloan, 2017). Initially discovered in mitochondria and later in chloroplasts, RNA editing in plants predominantly involves cytidine to uridine conversions (Giege & Brennicke, 1999; Figure 1). For example, Arabidopsis thaliana has over 500 mitochondrial and 34 chloroplast editing sites, mostly on transcripts encoding subunits of organellar protein complexes such as the NADH:ubiquinone oxidoreductase (mitochondrial complex I) or the mitochondrial ribosome (Bentolila et al., 2013; Hammani et al., 2009).
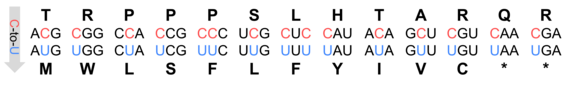
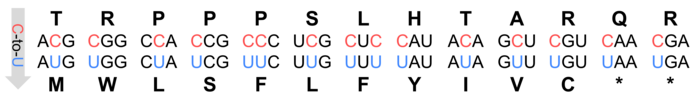
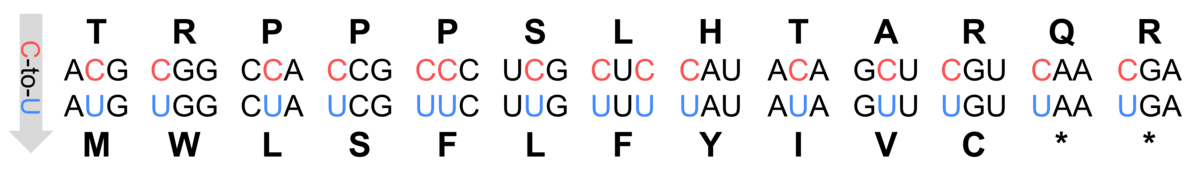
Figure 1: Codon changes by C-to-U RNA editing in Arabidopsis thaliana.
Plants employ numerous specificity factors, primarily pentatricopeptide repeat (PPR) proteins, to ensure precise RNA editing. These proteins, alongside multiple organellar RNA editing factor (MORF) proteins and other non-PPR helpers, form editosomes that conduct the editing process (Sun et al., 2016). While the mechanisms are well-studied, the purpose of RNA editing is debated. Some posit it's for restoring amino acids altered by DNA mutations (Gualberto et al., 1989), as these changes often enhance protein homology with other species (Sloan, 2017). Others suggest a regulatory role, indicated by variable editing frequencies across tissues and conditions (Bentolila et al., 2013; Zehrmann et al., 2008; Zeng et al., 2024).
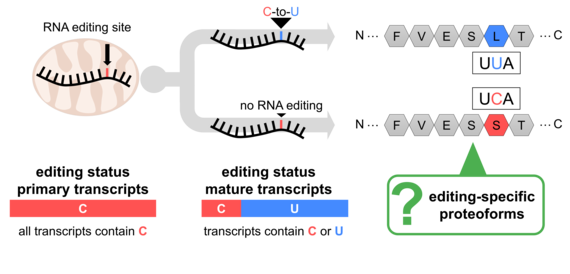
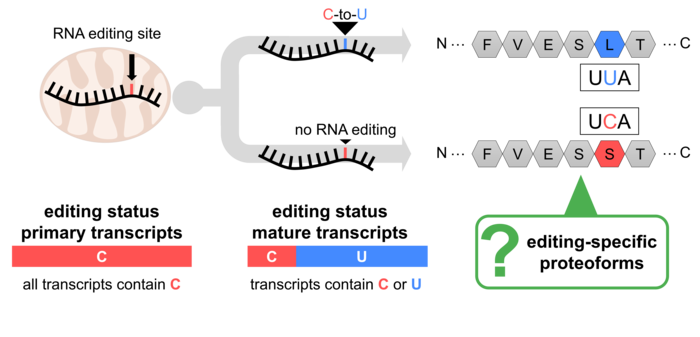
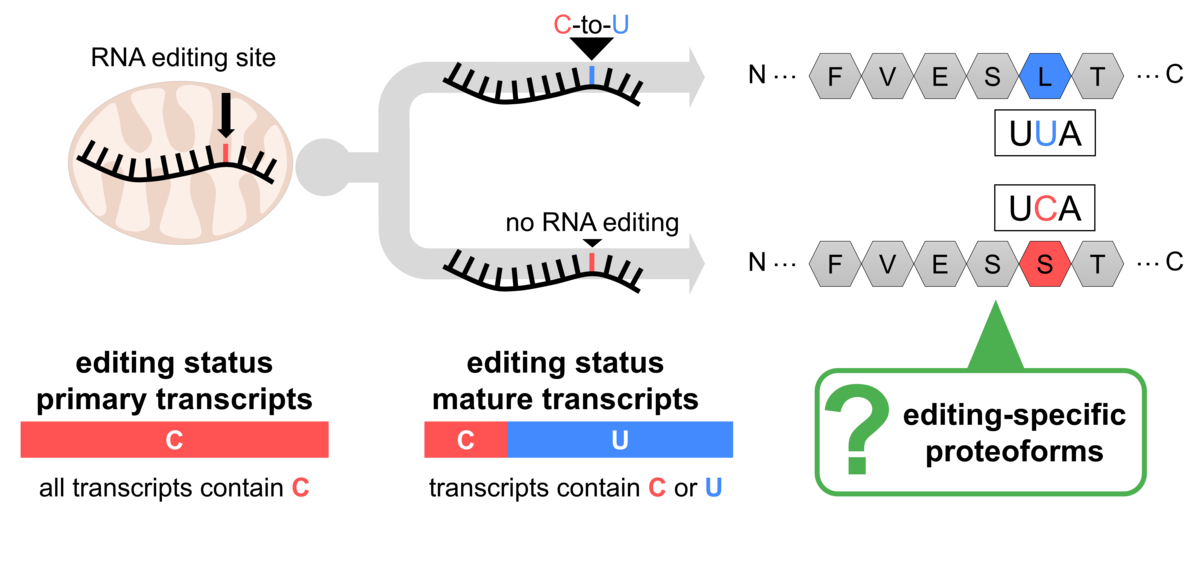
Figure 2: Partial RNA editing has the potential to diversify the plant mitochondrial proteome. Expression and localization of the respective proteoforms have not been studied so far.
This variability could lead to the synthesis of respective proteoforms termed "partially edited proteins," contributing to protein diversity (Figure 2). However, systematic studies of RNA editing's impact on the mitochondrial proteome are limited. Our recent project using "deep proteomics" aimed to address this gap and demonstrated that partially edited proteoforms are not only expressed but also incorporated into their protein complexes (Rugen et al., 2024; Figure 3).
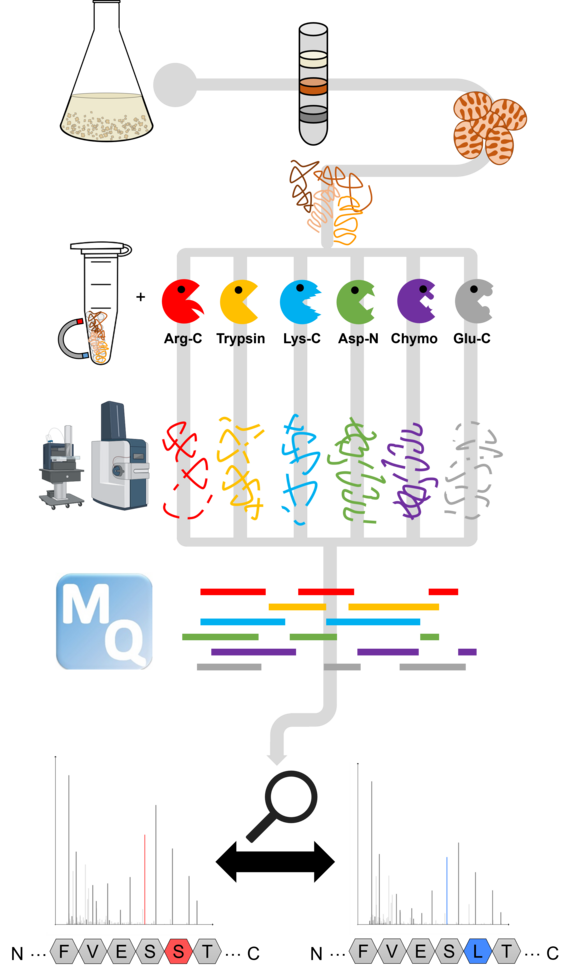

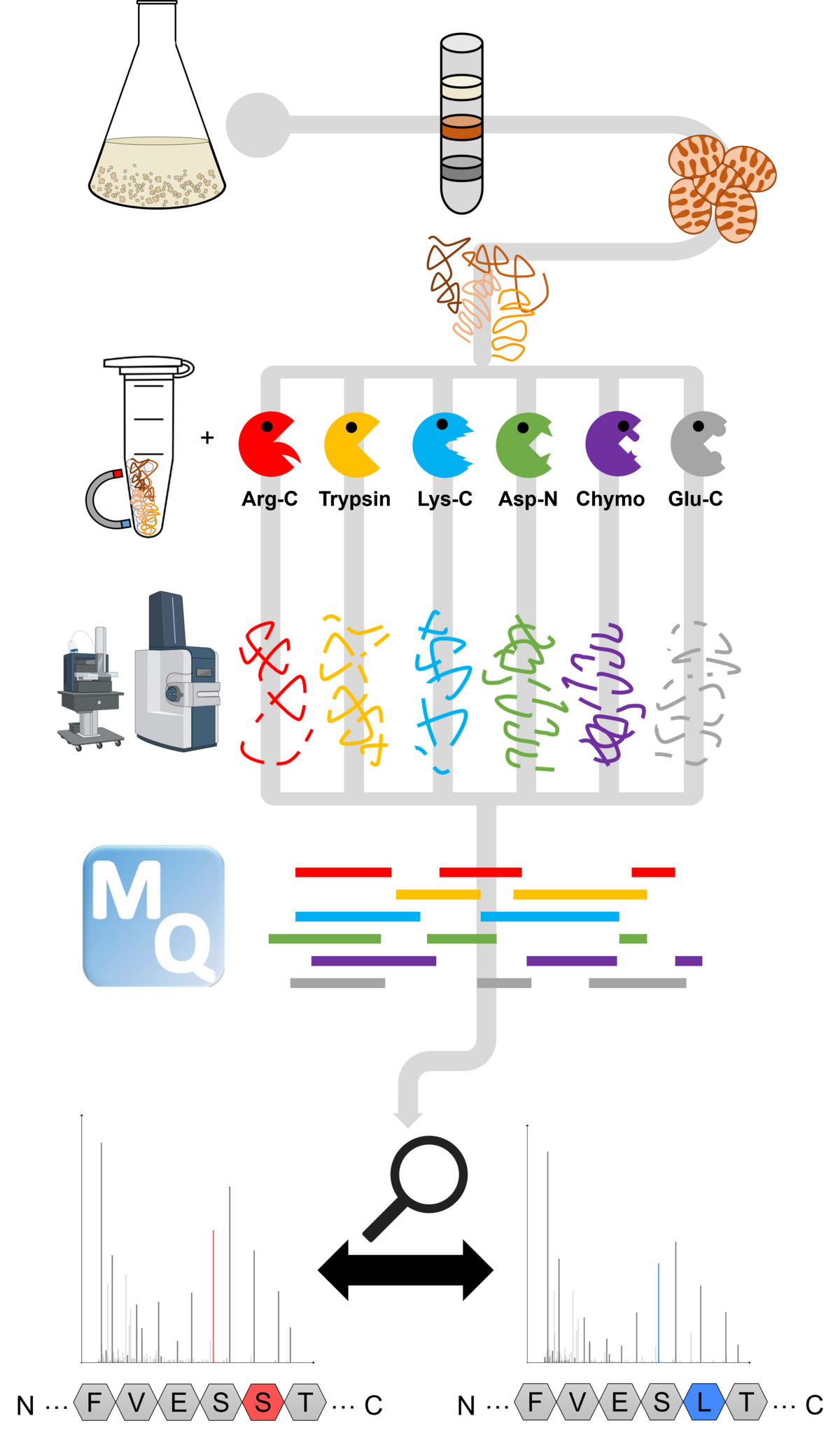
Figure 3: Multi-protease workflow of our recent deep proteomics investigation of the Arabidopsis thaliana mitochondrial proteome.
To better understand RNA editing, our group focuses on examining it from the protein perspective. We use advanced proteomic methods to detect partially edited proteins and track their fate within the organelles. Our core methods include comprehensive mapping of organellar proteomes using deep proteomics (Figure 3) and analyzing the composition of organellar protein complexes through complexome profiling (Figure 4). In this context, we primarily work with Arabidopsis thaliana and various RNA editing mutants.
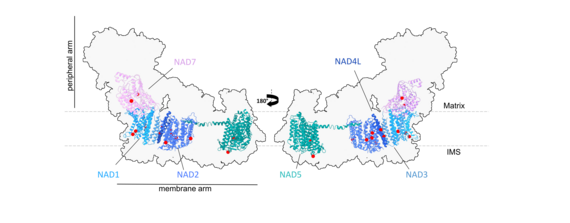
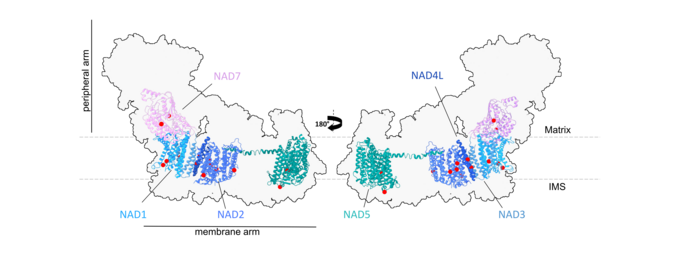
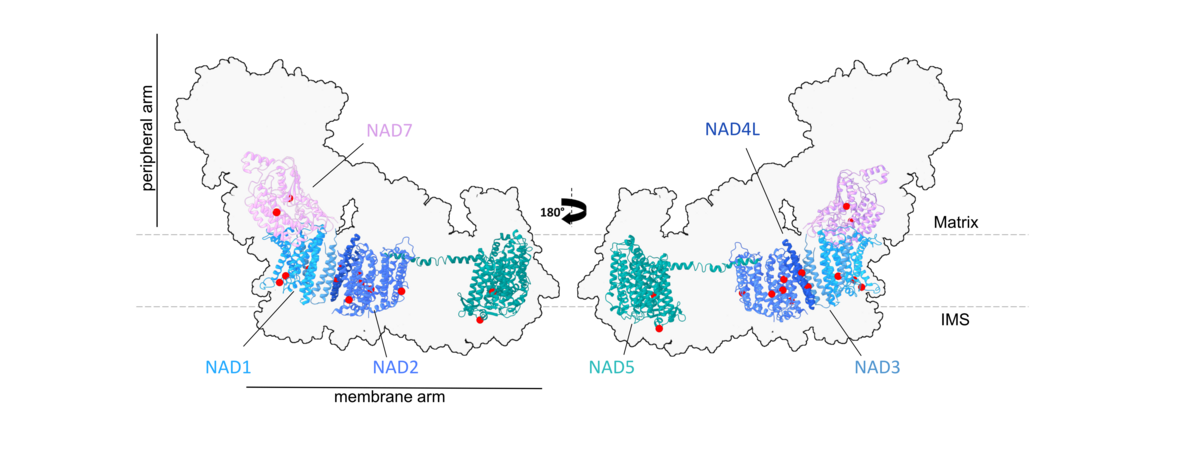
Figure 4: We aim to use modern proteomic approaches to characterize the RNA editing state of selected sites in mitochondrial Complex I and other protein complexes of Arabidopsis thaliana, where amino acid identity can be altered by RNA editing.


30419 Hannover


Publications cited
- Bentolila, S., Oh, J., Hanson, M. R., & Bukowski, R. (2013). Comprehensive High-Resolution Analysis of the Role of an Arabidopsis Gene Family in RNA Editing. PLoS Genetics, 9(6). doi.org/10.1371/journal.pgen.1003584
- Giege, P., & Brennicke, A. (1999). RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proceedings of the National Academy of Sciences of the United States of America, 96(26), 15324–15329. doi.org/10.1073/pnas.96.26.15324
- Gualberto, J. M., Lamattina, L., Bonnard, G., Weil, J. H., & Grienenberger, J. M. (1989). RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature, 341(6243), 660–662. doi.org/10.1038/341660a0
- Hammani, K., Okuda, K., Tanz, S. K., Chateigner-Boutin, A. L., Shikanai, T., & Small, I. (2009). A Study of New Arabidopsis Chloroplast RNA Editing Mutants Reveals General Features of Editing Factors and Their Target Sites. The Plant Cell, 21(11), 3686–3699. doi.org/10.1105/TPC.109.071472
- Rugen, N., Senkler, M., & Braun, H. P. (2024). Deep proteomics reveals incorporation of unedited proteins into mitochondrial protein complexes in Arabidopsis. Plant Physiology, 195(2), 1180–1199. https://doi.org/10.1093/plphys/kiad655
- Sloan, D. B. (2017). Nuclear and mitochondrial RNA editing systems have opposite effects on protein diversity. Biology Letters, 13(8). doi.org/10.1098/rsbl.2017.0314
- Sun, T., Bentolila, S., & Hanson, M. R. (2016). The Unexpected Diversity of Plant Organelle RNA Editosomes. In Trends in Plant Science (Vol. 21, Issue 11, pp. 962–973). Elsevier Current Trends. doi.org/10.1016/j.tplants.2016.07.005
- Zehrmann, A., van der Merwe, J. A., Verbitskiy, D., Brennicke, A., & Takenaka, M. (2008). Seven large variations in the extent of RNA editing in plant mitochondria between three ecotypes of Arabidopsis thaliana. Mitochondrion, 8(4), 319–327. doi.org/10.1016/J.MITO.2008.07.003
- Zeng, Y., Dong, J., Fu, D., Shi, M., Zheng, Z., Zhong, M., Wang, H. Bin, Duan, S. J., & Jin, H. L. (2024). The HPE1 RNA-binding protein modulates chloroplast RNA editing to promote photosynthesis under cold stress in Arabidopsis. FEBS Letters. doi.org/10.1002/1873-3468.14969


